*This course has been retired. There is no replacement course at this time. Please click here to view the current ATrain course listings.
To successfully infect a person, the influenza virus must develop ways to evade a person’s immune system. Viruses do this through evolutionary processes called antigenic drift and antigenic shift. Influenza type A viruses undergo both kinds of changes, while influenza type B and C viruses change only by the gradual process of antigenic drift.
Antigenic Drift: Continual Small Changes
Antigenic drift involves continual small changes or mutations to a virus’s surface antigens (HA or NA). Think of a small boat drifting across the ocean or clouds drifting across the sky. These changes produce new viral strains that are fairly closely related to one another and may be recognized by the immune system (sometimes called “cross-protection”). Changes due to antigenic drift can nevertheless accumulate over time, straining the ability of a person’s immune system to recognize the new virus.

Like clouds drifting across the sky, antigenic drift involves small, continual changes to a virus’s surface antigens. Source: Wikipedia Commons.
In most years, one or two of the virus strains in the influenza vaccine are updated to keep up with the changes in the circulating flu viruses. Changes in viruses due to antigenic drift can cause widespread infection because the protection that remains from past exposures to similar viruses is incomplete. Drift occurs in all three types of influenza virus (A, B, C).
Antigenic Shift, A Major Abrupt Change

Like this lightning storm near New Boston, Texas, antigenic shift involves major, abrupt changes in surface antigens (HA or NA). Source: Griffinstorm, Wikipedia Commons.
Antigenic shift is a major, abrupt change in one or both surface antigens (HA or NA). Shift occurs at varying intervals and likely is the result of reassortment (the exchange of a gene segment) between influenza A viruses, usually those that affect humans and birds.
Antigenic shift results in a new influenza A subtype that is so different from previous subtypes in humans that most people do not have immunity to the new virus. An antigenic shift can lead to a worldwide pandemic if the virus is efficiently transmitted from person to person.
An example of a “shift” occurred in the spring of 2009, when a novel H1N1 virus with a new combination of genes (from American pigs, Eurasian pigs, birds, and humans) emerged in people and quickly spread, causing a pandemic. Since the late nineteenth century, four occurrences of antigenic shift have led to major influenza pandemics.
Antigenic Drift and Shift of Influenza Strains
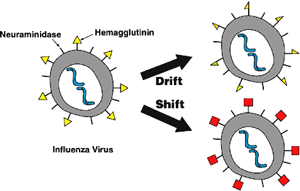
Antigenic drift vs. shift. Antigenic drift creates influenza viruses with slightly modified antigens, while antigenic shift generates viruses with entirely new antigens (shown in red). Source: Wikipedia Commons and USDA.
Although influenza viruses constantly and gradually change by antigenic drift, antigenic shift happens only occasionally. When a type A virus undergoes both kinds of changes, it is capable of evading host immunity, with profound implications for epidemiology and control. This is the main reason why seasonal influenza vaccines are updated frequently, to maintain protection in risk groups against currently circulating strains (Arinaminpathy & Grenfell, 2010).
Influenza Virus: Antigenic Changes | |
|---|---|
Antigenic drift |
|
Antigenic shift |
|
Influenza: Get the (Antigenic) Drift [2:52]
Source: NIAID. https://www.youtube.com/watch?v=ug-M1nIhfIA
The 2009 H1N1 Flu Pandemic—Quadruple Reassortment
The 2009 influenza A (H1N1) virus was a new flu virus that caused illness worldwide in March and April of 2009. This virus was originally referred to as “swine flu” because laboratory testing showed that many of the genes in this new virus were very similar to influenza viruses that normally occur in pigs in North America (CDC, 2010).
Further study showed that this new virus was different from the one that formerly circulated in North American pigs. It has two genes from flu viruses that have circulated in pigs in Europe and Asia, plus bird (avian) genes and human genes. Scientists call this a quadruple reassortant virus (CDC, 2010).
