Human influenza A and B viruses cause seasonal epidemics of disease (known as flu season) almost every winter in the United States. Influenza A viruses are the only influenza viruses known to cause flu pandemics, i.e., global epidemics of flu disease.
CDC, 2021
Types of Influenza Viruses
It is helpful to understand a little bit about the influenza virus—the different types, how they are named, and how they mutate. The more you know, the better you will be able to protect your patients, friends, and family members from catching the flu.
Influenza is a clever virus—it shifts, drifts, and adapts, every so often mutating into a virus to which humans have little or no immunity. When this happens, a pandemic can occur. The 1918 influenza pandemic killed tens of millions of people throughout the world; some of us lost grandparents, aunts, or uncles to the pandemic. All influenza A pandemics since that time, and almost all cases of influenza A worldwide, have been caused by descendants of the 1918 H1N1 virus (Taubenberger & Morens, 2006).
Influenza viruses are categorized and named by type. There are four types of influenza viruses—A, B, C, and D. Type is determined by the material within the nucleus of the virus.
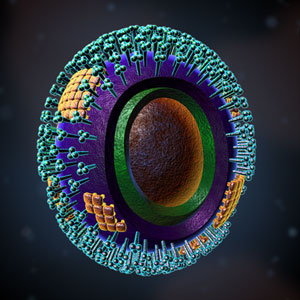
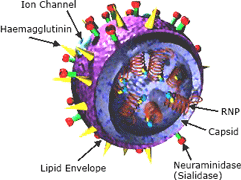
Left: The influenza virus. Copyright Zygote Media Group. Used with Permission. Right: Structure of the influenza virion. The hemagglutinin (HA) and neuraminidase (NA) proteins are shown on the surface of the particle. The viral RNAs that make up the genome are shown as red coils inside the particle and bound to ribonuclear proteins (RNPs). Source: NIH, public domain.
The nomenclature used to describe a specific influenza virus was established by the World Health Organization in 1980 and is expressed in this order:
- Virus type,
- Geographic site where the virus was first isolated,
- Strain or lineage number,
- Year of isolation, and
- Virus protein antigen subtype described by letter and number, H1 to H18 and N1 to N11.
For example, the 2009 H1N1 pandemic influenza virus was named as follows:
A/California/04/2009(H1N1)
This is translated as: Influenza type A, isolated first in California, lineage (strain) number 04, year 2009, and type H1N1.
In another example, a Fujian influenza virus that circulated in 2002 was named as follows:
A/Fujian/411/2002(H3N2)
This is translated as: Influenza type A, first isolated in Fujian (a province on the Southeast coast of mainland China), lineage (strain) number 411, year 2002, type H3N2.
The Fujian H3N2 influenza of 2002 caused an unusually severe 2003–2004 flu season, partly because it spread rapidly and partly because the vaccine for that season had already been formulated when the Fujian H3N2 virus was identified.
Influenza Virus Nomenclature
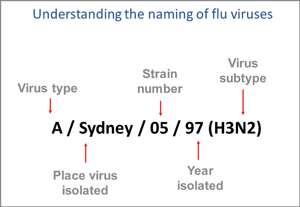
This image shows how influenza viruses are named. The name starts with the virus type, followed by the place the virus was isolated, followed by the virus strain number (often a sample identifier), the year isolated, and finally, the virus subtype. Source: CDC.
Type A Influenza and Its Subtypes
Type A influenza viruses are divided into subtypes, based on the presence of two glycoproteins on the surface of the virus. These glycoproteins are called hemagglutinin (HA) and neuraminidase (NA). About 18 hemagglutinins have been identified, although generally, only H1, H2, and H3 are found in human influenza viruses. There are more than 100 types of neuraminidase, but only N1 and N2 have been positively linked to influenza epidemics in humans.
While more than 130 influenza A subtype combinations have been identified in nature, primarily from wild birds, there are potentially many more influenza A subtype combinations given the propensity for virus reassortment* (CDC, 2021, November 2).
*Reassortment: the process by which influenza viruses swap gene segments. This genetic exchange is possible due to the segmented nature of the viral genome and occurs when two differing influenza viruses co-infect a cell.
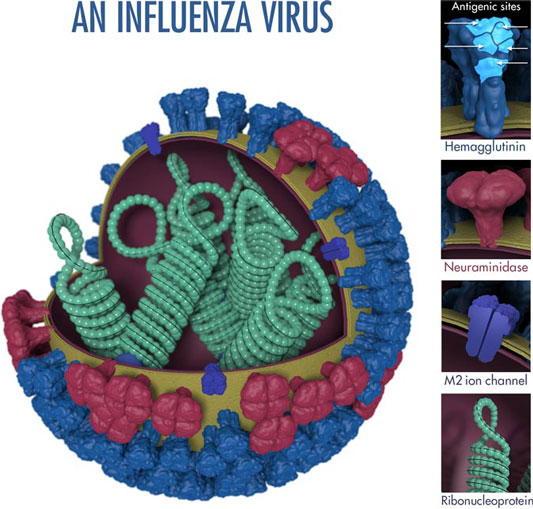
The above image shows the features of an influenza virus, including the surface proteins hemagglutinin (HA) and neuraminidase (NA). Following influenza infection or receipt of the influenza vaccine, the body’s immune system develops antibodies that recognize and bind to &dquo;antigenic sites,” which are regions found on an influenza virus’s surface proteins. By binding to these antigenic sites, antibodies neutralize flu viruses and prevent them from causing further infection. Source: CDC.
Hemagglutinin and neuraminidase are also called antigens, substances that, when introduced into the body, stimulate the production of an antibody. Currently, there are two subtypes of influenza A viruses found circulating among human populations: influenza A (H1N1) and influenza A (H3N2).
Wild Birds Provide the Usual Reservoirs
A reservoir is the place where a pathogen lives and survives. Wild aquatic birds—particularly certain wild ducks, geese, swans, gulls, shorebirds, and terns—are the natural hosts for most influenza type A viruses (CDC, 2017).
Most influenza viruses cause asymptomatic or mild infection in birds; however, clinical signs in birds vary greatly depending on the virus. Infection with certain avian influenza A viruses (for example, some H5 and H7 viruses) can cause widespread, severe disease and death among some species of birds (CDC, 2017).
Type A Influenza Viruses Also Circulate in Pigs
Pigs are susceptible to avian, human, and swine flu viruses and can potentially be infected with influenza viruses from different species. If this happens, it is possible for the genes of these viruses to mix (reassort) and create a new virus.
Influenza viruses that normally circulate in pigs are called “variant” viruses when they are found in people and denoted with a letter “v.” H3N2v viruses from the 2009 H1N1 pandemic virus were first detected in people in 2011 and were responsible for a multi-state outbreak in the summer of 2012 that resulted in 306 cases, including 16 hospitalizations and 1 fatality (CDC, 2019, January 3).
Most cases of H3N2v identified during 2012 were associated with exposure to pigs at agricultural fairs. Many fairs have swine barns where pigs from different places come in close contact with each other and with people. These venues can allow the spread of influenza viruses both among pigs and between pigs and people. Just as in humans, infected pigs can spread influenza viruses even if they are not symptomatic. Although instances of limited person-to-person spread of this virus have been identified in the past, sustained or community-wide transmission of H3N2v has not occurred (CDC, 2019, January 3).
Type B Influenza
Influenza type B viruses are separated into two genetic lineages (B/Yamagata and B/Victoria). They are not classified by subtype like influenza A viruses. Influenza B viruses from both the Yamagata and Victoria lineages have co-circulated in most recent influenza seasons, a major factor in adding 2 influenza B viruses to the 2021-2022 flu vaccine. The quadrivalent vaccine for 2021–2022 contains both the Yamagata and Victoria lineages (CDC, 2021, November 2).
Influenza type B viruses are usually found only in humans but in general are associated with less severe epidemics than influenza A viruses. Although influenza type B viruses can cause human epidemics, they have not caused pandemics. This is thought to be because influenza B viruses undergo genetic changes less rapidly than influenza A viruses.
In a review of laboratory-confirmed influenza cases from 2001 to 2018, data from the CDC and several European countries indicated that influenza B has been a major contributor to the total morbidity and mortality from influenza. During that time, 15% of all influenza-attributable respiratory and circulatory-related death in the U.S. and 34% among pediatric patients were attributed to influenza B. A recent study from Israel found similar rates of ICU admission and disease severity in patients hospitalized with either influenza A or B (Shasha et al., 2020).
Type C Influenza
Influenza C, a lesser-known type of influenza commonly causes cold-like symptoms and sometimes lower respiratory infection, especially in children <2 years of age. It is mainly a human pathogen; however, the virus has been detected in pigs, dogs, and cattle, and rare swine–human transmission has been reported. Seropositivity* has been found to be as high as 90% by 7–10 years of age, suggesting that most people are exposed to influenza C virus at least once during childhood (Sederdahl and Williams, 2020).
*Seropositivity: having a positive test result for the presence of a specific antibody in the serum of the blood.
Influenza type C is less common and less studied than influenza A and B. The virus lacks the multiple subtypes (hemagglutinin and neuraminidase) found in influenza A, which limits its ability to mutate. Influenza C is thought to be unlikely to cause a pandemic, although localized epidemics have occurred. As with type B influenza viruses, type C influenza viruses are not classified according to subtype.
Type D Influenza
A new type of influenza virus, known as type D, has recently been identified in cattle and pigs. Influenza D virus infection in cattle is typically asymptomatic; however, its infection in swine can cause clinical disease. Swine can also be infected with all other types of influenza viruses, namely A, B, and C. Consequently, swine can serve as a “mixing vessel” for highly pathogenic influenza viruses. People working closely with calves have higher rates of seropositivity to the influenza virus (94%) than the general population (1.3%) (Kesinger et al., 2018).
Types of Influenza Virus |
|
|---|---|
Influenza type |
Characteristics |
Type A |
|
Type B |
|
Type C |
|
Type D |
|
